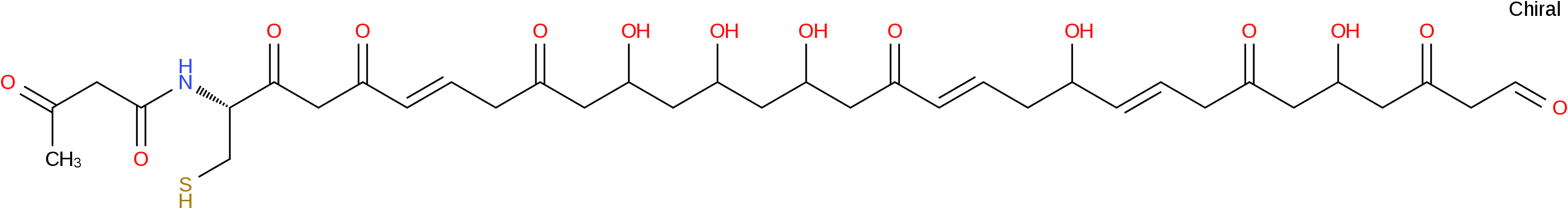
Gene cluster description NZ_ADVE02000002 - Gene Cluster 1. Type = arylpolyene-transatpks-nrps. Location: 70411 - 175756 nt. Click on genes for more information.
arylpolyene: (APE_KS1 or APE_KS2)
Homologous gene clusters All hits Methylosinus trichosporium OB3b ctg00105, whole genome sho... (100% of genes show similarity) Leptolyngbya sp. PCC 7376, complete genome. (41% of genes show similarity) Burkholderia thailandensis MSMB59 chromosome 2, complete sequ... (50% of genes show similarity) Brevibacillus brevis FJAT-0809-GLX JK200063, whole geno... (45% of genes show similarity) Burkholderia thailandensis E254 chromosome 2, complete sequence. (45% of genes show similarity) Burkholderia thailandensis USAMRU Malaysia #20 chromosome 2, c... (41% of genes show similarity) Burkholderia gladioli strain DMSZ11318 putative regulator gene... (37% of genes show similarity) Bacillus subtilis subsp. subtilis str. 168 complete genome. (41% of genes show similarity) Bacillus subtilis subsp. subtilis str. 168, complete genome. (41% of genes show similarity) Bacillus subtilis subsp. subtilis str. AG1839, complete genome. (41% of genes show similarity) Download graphic
Homologous known gene clusters All hits Oocydin A biosynthetic gene cluster (21% of genes show similarity) Oocydin A biosynthetic gene cluster (21% of genes show similarity) Kalimantacin / batumin biosynthetic gene cluster (17% of genes show similarity) Bongkrekic acid biosynthetic gene cluster (17% of genes show similarity) Mupirocin biosynthetic gene cluster (13% of genes show similarity) Thailandamide biosynthetic gene cluster (13% of genes show similarity) Bacillaene biosynthetic gene cluster (13% of genes show similarity) Difficidin biosynthetic gene cluster (8% of genes show similarity) Corallopyronin biosynthetic gene cluster (8% of genes show similarity) Calyculin biosynthetic gene cluster (8% of genes show similarity) Download graphic